This is a quick follow-up to Making RMarkdown the New Way You Write Everything, which among other topics, described how to write your article manuscripts in RMarkdown. The assumption there was that you’d want a nice PDF as output to make sharing your work easy. But what if you need to quickly get a word-formatted version of your article? Now you may be asking yourself, “Why in the name of all that’s good would I ever want that?”
The Problem
We are in the process of preparing your article for publication. We have noticed that manuscript and the files supplied are in PDF format, and we cannot typeset your article from a PDF file. We would be most grateful if you could re-supply the files for your manuscript within 24 hours by e-mail.
Our preferred format for the manuscript and tables is Word. Optimal resolution for line artwork is 800 dpi and for halftones is 300 dpi. Preferred formats are EPS and TIFF for line artwork, and JPEG for halftones.
Ugh. Right. That’s why.
The Solution
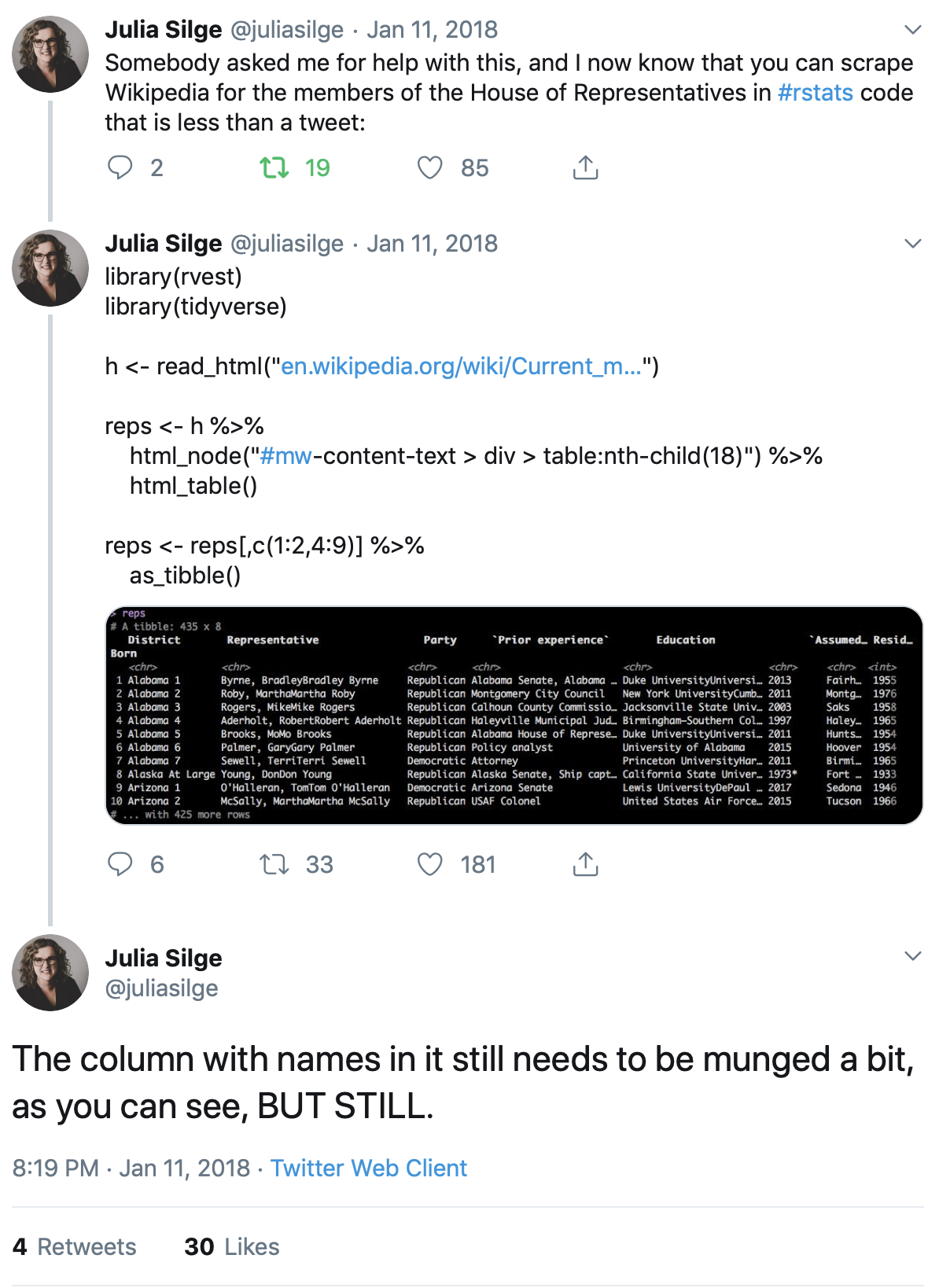
RMarkdown makes converting a manuscript to a .docx fairly easy. The process is not quite seamless, though, so it’s wise I think to duplicate the .Rmd file and rename it with _word. That is, if the paper starts from example_article.Rmd, make a duplicate and call it example_article_word.Rmd.
Step 1. The YAML metadata
Our preferred PDF template, svm-latex-ms2.tex, has all kinds of YAML stuff that the word version will ignore, although it doesn’t actually have to be deleted: fontsize, spacing, citecolor, linkcolor, endnote.1 But author apparently has to be a single string—author: "Frederick Solt, _University of Iowa_"— rather than using name and affiliation, and thanks should be switched to an inline footnote at the end of the title:
title: "An Example Article^[This paper's revision history and the materials needed to reproduce its analyses can be found on Github at <http://github.com/fsolt/example_article>. Corresponding author: [frederick-solt@uiowa.edu](mailto:frederick-solt@uiowa.edu).]"The big thing to change, of course, is the output, which should appear as below:
output:
word_document:
reference_docx: word-styles-reference-01.docx
keep_md: trueThe word-styles-reference-01.docx file ensures the references get hanging indents; one could do more here to customize the format, but we don’t care, and even this much probably isn’t really necessary. Anyway, download it here and put it in the same directory with the .Rmd.
Setting keep_md to TRUE has the side effect that the directory of figures is preserved. They’ll all need to be exported as .tiff and .jpeg files, too (let the publisher figure out which format they want to use for each, I think). I’m positive there’s a clever one-liner to do this, and I really want to go look for it, but it took way less than a minute to get the job done in Preview. I’m sure there are equally quick ways to do this in windows. Satisficing, FTW.
Oh, one more thing—my little hack to find the bibliography automatically
bibliography: \dummy{ `r file.path(getwd(), list.files(getwd(), "bib$"))` }doesn’t seem to mix well with output: word_document. Just call the .bib file directly, e.g., bibliography: "example_article.bib".
Step 2. Figure Captions and Cross-References
The only in-text issue I found is that figure captions don’t have the “Figure X:” prefix and figure cross-references don’t appear. So go into the .Rmd, search for “fig”, and add “Figure 1:”, “Figure 2:” and so on to the figure captions in the code chunks, and add the “1”, “2”, etc., where they go in the text.
Step 3: Bibliography
There are two things to do here. First, add “## References” to the bottom of the .Rmd. Second, carefully look over the output bibliography to catch any differences between how your references are cited in the PDF and how they appear in the word file. (In my case, my .bib includes many unpublished documents that get cited how I want them to in the PDF, but don’t appear right in the .docx; they need to be switched to misc to appear correctly. For each of these items, in my favorite Bib\(\TeX\) software, BibDesk, I needed to cut the Note entry to the clipboard, switch the item to misc, and paste the clipboard into Howpublished.)
So, if any such modifications are necessary, one should create a new, duplicate version of the .bib file with the _word name suffix—remember to change this name in the bibliography: entry of the YAML header of the .Rmd, too—and make the needed changes in that file.
Done!
Not instantaneous, but pretty darn quick, really: a matter of minutes, now that it is all figured out.
I delete it anyway, because I prefer my code clean. You of course can feel free to do you.↩︎